AKos Consulting
& Solutions Deutschland GmbH
Telephone
+49 7627 970068
Fax
+49 7627 970067
Fax to Mail
+49 1805 744743 8318
E-Mail
General
Information:
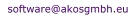
|
|
Confirm or
reject future newsletters
Our mailing
list is generated mainly from business correspondence. We use
automatic procedures to do this.
Please send an email to
subscribe@akosgmbh.de if you want to receive future newsletter,
or send an email to
unsubscribe@akosgmbh.de if you want to be removed from the
mailing list.
Please
forward this to a friend, who might be interested.
|
|

CWM Global Search allows searching the Internet by
structure, synonym and CAS Registry Number. In a Quick
Search, we return structure, names and
CAS Registry Numbers* within seconds. The Global Search
allows a comprehensive search and the resulting links are
organized by topics. A search by name can automatically
invoke another search by structure and/or CAS Registry
Number, or
any combination of these. CWM Global Search presently
searches 50 free chemical and pharma relevant databases -- containing
more than 100 million pages which associate chemical
structures with data.
All you have to do is to open
the URL
http://cwmglobalsearch.com/gsweb
in your Internet Explorer or Firefox on Windows, or Safari on Macintosh.
Please let us
know of sources of your interest and we will try to include them.
|
Name to Structure
Type a valid systematic chemical name in
the associated textbox.
A click on the 'Name to Structure' button tries to convert the chemical
name to a structure.
If the conversion was successful, the structure box of the QuickSearch
page will be updated with the generated structure.
Chemical Name Search
We explained the last time how to use this
option for substructure search by name. This option also helps if you
have an IUPC name that has an error. Leave away the brackets search by
fragments and the system suggest structures that match the fragments in
the name.
New features:
- Quick Search starts automatically if
user press ENTER key after entering name or CAS Registry Number
query
- Pressing ENTER key after
entering/editing Names/CAS Registry Numbers in GS query page,
automatically populates name/CAS Registry Number listbox
- Visible indication of used
‘Predefined profiles’
- Possibility to Save/Load QuickSearch
and Global Search queries
- New datasource: EBI search
- If more than one query term is
entered in the text search field we create automatically two
queries, one with quotation mark, and one without. This reminds the
user if he wants to search by "or" or by "and" logic.
- The Quick Search has two extra
options, the "Chemical Name Search", and "Tautomerize structure
query".
- Generation of IUPAC name for
chemical structures in the QuickSearch page using ChemAxon's
Structure to Name service.
- Structure boxes are now
automatically resized.
- New added horizontal and vertical
splitters makes it easier to use CWM Global Search on devices with
small screens.
- Support for chemical structure
searches in Wikipedia.
|
At the first time you
will be asked to install the Microsoft Silverlight Plug-In, provided it
is not already installed.
We recommend to start by
reading the installation notes at
cwmglobalsearch.com/installation.
A free version is
available that is restricted to searches at Google, PubChem, ChemSpider
and AKosSamples.
The full version is
240 Euro per user per year.
We sincerely are
interested in your feed-back. Please use the feedback button in the
application. |
|
Electronic
Lab Notebook
affordable for everyone!
iLaaber - the cloud-based ELN
With only a few clicks you
can launch a scientific workspace, invite your colleagues
and collaborators and gather all your experiments in one
secure and searchable location. With iLabber you do not need
to hire costly consultants nor purchase any additional
hardware. With iLabber you can start your first ELN
experiment in five minutes...
|
|
 |
|

Spot the difference -
and get it right!
Can you name proteins just by looking at them? Proteax DerNot
expressions clearly show you how much two proteins differ. Works with
cyclic chains too.
Protein SAR tables - impossible you say ? No, not at all.
Proteax can name your biopharmaceutical candidates so you get consistent
and meaningful labels for your protein SAR tables.
Having trouble with your weight ?
Does the analytical department claim a molecule weight of 9243.5 Da but
your sequence says 9202 Da ? Let Proteax enable seamless communication
between mass-spec and medicinal chemistry.
Can your chemists and bioinformatics people agree ?
Is it a structure ? Is it a sequence ?... Imagine that they used Proteax
- a consistent way of representing biopharmaceuticals that lets you see
molecules as sequences or structures as you please.
"Can you generate 1000 controlled mutations for me this afternoon ?"
Applied DerNot expressions let you create protein derivatives through
simple Excel expressions. With chemically consistent results. |
Proteax®
for Spreadsheets 1.3 released
Proteax for Spreadsheets 1.3 is now ready for download. Improved layout
and graphical rendering of chemical structures.
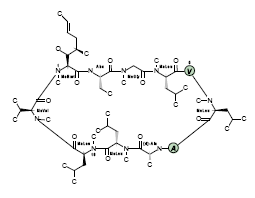
- The image to the right is an actual rendering of Cyclosporin CsA
generated directly from sequence. Unmodified natural amino acids are
shown as green "bubbles".
- Optional name and number residue labels.
- 64-bit Microsoft Excel® 2010 directly supported.
You can get a fully functional trial version of Proteax for Spreadsheets
by downloading it here.
|
 |
Proteax Desktop and Proteax Cartridge downloads
With the release of Proteax 1.3 all Proteax products will be available
for direct download
from the Biochemfusion web site.
- Proteax Desktop - Use Proteax from .NET, Python, Pascal, or
C/C++.
- Proteax Cartridge - Use Proteax SQL extensions and indexing
inside Oracle® databases.
Discounted licenses until year-end
There is still time to get a Proteax for Spreadsheets license for only €
350,-. This is an introductory offer that expires December 31st. |
Can your chemists and
bioinformatics people agree ?
Is it a structure ? Is it a sequence ?... Imagine that they used
Proteax - a consistent way of representing biopharmaceuticals that
lets you see molecules as sequences or structures as you please.
"Can you generate 1000 controlled mutations for me this afternoon
?"
Applied DerNot expressions let you create protein derivatives
through simple Excel expressions. With chemically consistent results. |
|
Predicting Potential Adverse Drug
Reactions Caused by Off-Targets
Using AurPASS
Free Webinar
When ?
Thursday, June 30th
8:00 AM
PDT
11:00
AM
EDT
5:00 PM
Paris
Presenter
Ismail
IJJAALI

  
Product Development
|

Abstract
Adverse drug
reactions resulting from off-target activities account for the majority
of drug withdrawals from the market. With the credo 'fail early, fail
cheap',
in silico
methods are expected to gain largely in importance with improved models
that are able to address different aspects of drug safety at early
stages of drug discovery projects. These techniques consist of a
spectrum of methods designed to efficiently search large compound
databases
in silico
for molecules likely to have a desired biological activity or to avoid
unwanted off-target side effects.
In this
webinar, we will be introducing AurPASS (Aureus Prediction of Activity
Spectra of Substances). You will learn how this application exploits
high quality experimental Structure-Activity Relationship data combined
with PASS probabilistic ligand-based methodology to generate
in silico
biological activity profiles of new substances and therefore help gain
insights into potential off-targets.
Following the
presentation, this webinar session includes a demonstration of how
AurPASS generates
in silico
pharmacological profiles and helps investigate potential adverse effects
and related safety pharmacology issues.
What Will Be Covered
-
Generation of
in silico
biological activity profile(s) of your compound(s) on different
target families (GPCRs, Kinases, Ion Channels, Nuclear Receptors).
-
Probability
estimates of predicted activities for a better assessment of the
activity spectrum.
-
Detection of
potential off-target effects earlier in drug discovery process.
Who Should Attend
-
Medicinal
chemists and pharmacologists
-
Computational
chemists
-
Safety
pharmacologists
|
AurPASS®
(Aureus Prediction of Activity Spectra of Substances) is a software tool
which exploits high quality experimental SAR data to generate in
silico biological activity profiles of new substances.
AurPASS®
is a ready-to-use software tool running with several SAR bases extracted
from the Aureus knowledge bases. These include GPCR, Kinase, Ion
Channel, Nuclear Receptor, Protease, and hERG.
AurPASS®
is the result of the combination of first-in-class systems and high
level expertise provides robust computer-assisted estimates with more
than 90% accuracy.
PASS (Prediction of Activity Spectra of Substances) predicts
Biological Activity Spectra of compounds.
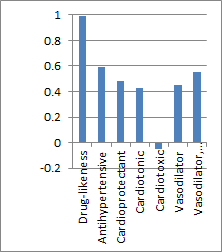
Biological Activity Spectrum of a
compound.
X-Axis = Pa-Pi
Y-Axis = Activities
Pa = probability that the compound has
this activity
Pi = probability that the compounds does not have this activity
Only a few of the more than 4000
activities are shown for which PASS is trained. |
|
We are very pleased to announce that the
latest release of our free
Discovery Studio
Visualizer software, version 3.0, is now available for
download at: http://accelrys.com/products/discovery-studio/visualization-download.php
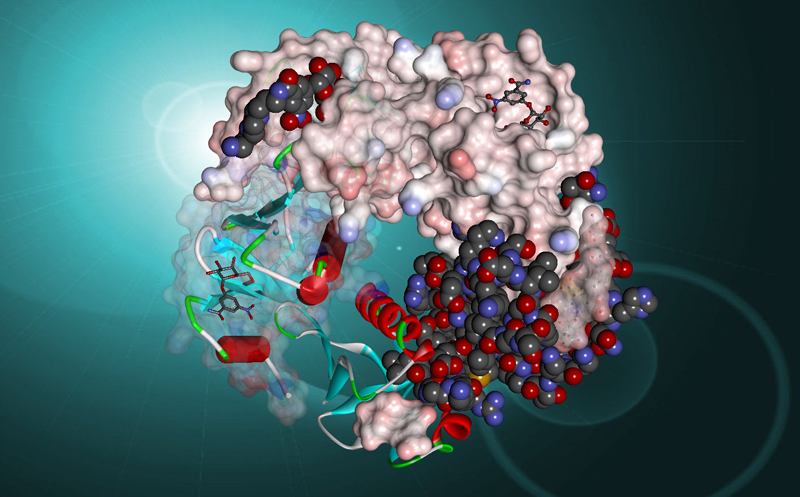
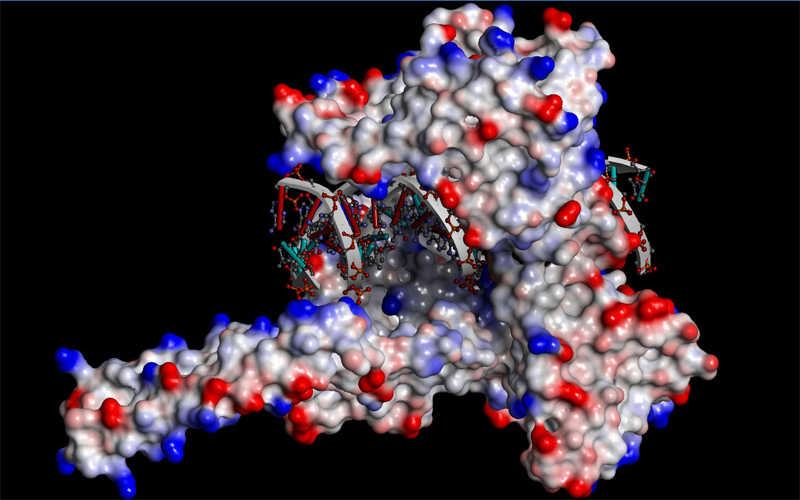
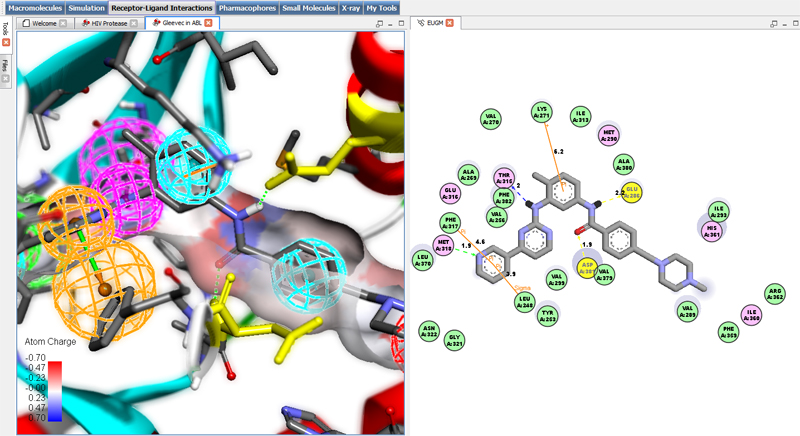 |
With Discovery Studio (DS) Visualizer,
the following features are available for free:
New with version
3.0:
All this is
available for free! To learn more about the visualization capabilities
in DS Visualizer 3.0 download our data sheet:
http://accelrys.com/products/datasheets/discovery-studio-visualizer-30.pdf .Download
DS Visualizer 3.0 today at:
http://accelrys.com/products/discovery-studio/visualization-download.php
Best regards,
Adrian Stevens (Product Marketing Manager, Discovery Studio) |
AKos GmbH
is the exclusive reseller for Accelrys in
the countries East of Austria until the Chinese border and South
including Macedonia and Serbia, with the exception of academia in Russia and Poland.
The products of Accelrys are:
|
|
Webinars by Accelrys: |
Quantifying the Value of
Pipeline Pilot, with Axios Partners.
The recording is available if you would like to review it at a time that
is convenient:
http://media.accelrys.com/webinars/pipeline-pilot-85/6-16-11-quantify-value-pp.wmv
You may also be interested in upcoming webinars in this series
Learn more and register here:
http://accelrys.com/events/webinars/pipeline-pilot-8/index.html |
|
|
European User Group Meeting
Oct 11-13, Athens
Greece
Academic registrants receive a 250 $ discount through the start of the
conference, but are encouraged to register today. Learn more by visiting
the
EUGM website.
|
 |