| |
Efficient Internet Searches for Chemists
Alexander Kos *, Hans-Jürgen Himmler
AKos Consulting & Solutions Deutschland GmbH (AKos GmbH), Austr.
26, D-79585 Steinen, Germany
* Author
to whom correspondence should be addressed; E-Mail:
software@akosgmbh.de;
Austr. 26, D-79585 Steinen,
Germany, Tel.: +49 7627 970068; Fax: +49 7627 970067.
Abstract:
iScienceSearch is a free Internet application that allows the user to
search by structure, synonyms, CAS Registry Numbers and free text over
100 databases on the Internet. Google is one of these databases. For
chemical structure related questions iScienceSearch is a better choice
than the Google front-end. Depending on the question sometimes a search
started in databases like PubChem or SciFinder is more suitable,
sometimes searching the Internet with iScienceSearch gives better
results.
Besides searching the Internet, iScienceSearch offers tools, like a
direct link to predict biological activities and toxicities. The
application can be started using the URL http://isciencesearch.com/iss.
Keywords:
Internet Search Engine, Meta-search engine, Rich Internet Application,
RIA, iScienceSearch, Chemical Structure Search
Introduction
Most people go to Google, if they want to know more about a subject [1],
[2]. Most chemists use PubChem, or SciFinder, if they
want to know more about a compound. Both of these are databases and not
Internet search engines. Is there no Internet search engine for
chemists? There is iScienceSearch [3].
Why would you use
iScienceSearch and not Google?
With iScienceSearch, you can search the Internet by chemical structure.
Sometime, if you search for a specific chemical name in Google, you get
no relevant answer at all.
iScienceSearch extends your query and searches not only by the
specific chemical name. If you start with a name, iScienceSearch will
find the CAS Registry Number [4], provided it is in
the public domain, the structure, and more names. Sometimes
iScienceSearch does more than 100 different searches in the background.
For instance, search for toxicity by structure and you will get a link
to a database, which only can be searched by CAS numbers. Search in
Google for plants that contain maslinic acid and you will never find the
Wikipedia page [5] for clove, because it mentions
only crategolic acid, since crategolic acid is a synonym for maslinic
acid.
Get only relevant answers. You can restrict the search to profiles.
Search in “Supplier” if you want to buy a compound. Search in “Open
access” journals if want to make sure that you do not only get an
abstract.
In Google you look at the first page, maybe you look at the second page.
This means sometimes you miss the most relevant answer. One of the
largest collection of screening compounds is AKosSamples [6].
If you search “buy research screening compounds” you will need to go to
page 3 in Google to find the link for AKosSamples. The result page in
iScienceSearch gives a different view. iScienceSearch groups according
to sources. For instance in a search for “Origins of life” PubMed [7]
obviously provides you a scientific and not a philosophical text. In
Wikipedia, you can expect both.
iScienceSearch gives you the most current view of the Internet. There is
always a time delay between publication and when the data is recorded in
a database. A structure published in PubChem [8]
will appear in about 14 days in Google, a structure published in
AKosSamples will appear about 4 weeks later in CHEMCATS [9],
and these are the short delays.
Google is a database [10], and as such a source in
iScienceSearch. If you know how to transform a chemical structure
drawing into InChI name or key [11] you could also
search Google by structure. iScienceSearch does this automatically for
you. However, you definitely cannot do a substructure search in Google.
How often do chemists miss a structure because they start with the enol
form and in the publication or database is only the keto form?
Google cannot index an Oracle [12], MySQL
[13] , etc. database. If the data are not in an html
file, or are server side generated asp/php etc. pages the data will not
appear in the Google index [14], [15].
For instance AKosSamples is a MySQL database and you need a special
interface to search the database. This problem does not exist in a
federated search if access to the database is provided. For examples as
it is for AKosSamples in iScienceSearch.
The heading to this paragraph was “Why would you use iScienceSearch and
not Google”. For chemical questions indeed it makes more sense to use
iScienceSearch instead of Google. In the following we compare
iScienceSearch to databases. Here it depends on your question if you
start with a database or iScienceSearch, or use both. For some searches
a database is the better choice. You can use Boolean logic in your
searches and restrict your searches to certain fields in the database.
Why would you use
iScienceSearch and PubChem?
PubChem is a database and there are time delays, see below. No system
can be comprehensive. Building a database with all suppliers is just too
expensive. For instance PubChem has 155 suppliers, CHEMCATS has ca. 880
[9], eMolecules [16] ca. 140,
ChemSpider [17] has in total 493 sources; ChemExper
[18] lists more than 1500 suppliers. Experience has
shown that iScienceSearch is the system of choice if you are searching
for suppliers of research chemicals, because with the exception of
CHEMCATS all these and 26 more directories of suppliers can be searched
in iScienceSearch in one go.
Why
would you use iScienceSearch and SciFinder?
The foremost reason to use
iScienceSearch is cost. iScienceSearch is free. With the exception of
CHEMCATS, the basis of the Chemical Abstract database are journals,
patents, dissertations and other high quality sources
[19], but not other databases like ChEMBL [20]
which collect also high quality data. It should be obvious that not
everything is in SciFinder. A few examples are at the end of the paper.
There is one more reason why a chemist should also search in
iScienceSearch, it is the “Extended Search”:
Extended Search
We chemist have solved the issue of similarity by using substructure,
and similarity searches with chemical structures. It is extremely
limiting that many databases on the Internet cannot be searched by
structure.
In iScienceSearch we implemented the extended search. This means when
you draw a structure, or type a chemical name, iScienceSearch searches
in the background databases and finds concordances of structure,
identification numbers (i.e. CAS Registry Number or AKos Number), and
names. For Aspirin you will find about 200 different names, and it would
be too time consuming to do 200 extra searches in the background.
iScienceSearch limits the names to about 20 most important ones. In the
background iScienceSearch searches for instance by different InChIs, CAS
Registry Number and names.
The result is that you start with a structure and get answers from a
database that can only be searched by CAS Registry Number (see list of
databases in the Appendix for examples), or you start with a name like
maslinic acid and get perfectly correct results where only the synonym
crategolic acid
appears.
Profiles
A profile is a selection of databases that are relevant for specific
searches. If you want to buy a compound, you can choose to search only
over databases that provide supplier information. In a federated search
over the Internet it is yet impossible to use a logical “and”. If the
original source can interpret a query like “pyridine and carcinogenic”,
you will get only answers where pyridine is connected with carcinogenic.
However, you cannot draw a structure and type carcinogenic and expect to
find only such structures that are carcinogenic. This would mean that
the system needs to collect all answers from the Internet, builds a
local cache (database) and filters the search. A profile helps to
overcome this limitation. If you want to find LD50ties, you search in
the profile “Toxicity” and search only over databases that hopefully
offer a LD50. However, an LD50 can always be mentioned in a journal
article. Then you should extend your search over the profile
“Literature”. Another strategy is to begin searching over “All Sources”
and use the sort, group, and filter methods in the result page.
Additional features
Some of the iScienceSearch tools fall in the category of predicting
data. iScienceSearch shows links to experimental data where possible.
Some features are convenient, like generating structure from text. Other
tools are there to compare results of the different databases, to
discover error and
discrepancies.
|
Feature/Tool |
Purpose |
Explanation |
|
Name to
structure |
You do not have to draw a structure! |
You can generate a structure by giving a name (IUPAC,
synonym), CAS #, AKosNumber, InChI, etc. |
|
Compare structures |
What is the right structure?
|
With a structure on the screen or a name, CAS #, etc. in the
text box you will get a grid comparing the structures as
they look in different databases. This is very useful to
check your structures before you publish, i.e. “Tracleer”
and “Bosentan”, see below. |
|
Compare activities |
What is the major activity of a compound? |
With a structure on the screen or a name, CAS #, etc. in the
text box you will get a grid comparing the activities as
reported in different databases. |
|
Predict chemial properties |
What is the correct melting point? |
With a structure on the screen or a name, CAS #, etc. in the
text box you will get a grid with GUSAR [21]
calculated physical properties and the links to calculated
properties by ACD Labs [22] and ChemAxon
[23]. |
|
Predict biological activities |
Which are the possible biological effects of a compound? |
With a structure on the screen you will get a reliable
prediction of effects, like toxicities, biological
activities, etc [24]. |
|
chemicalize |
What is the IUPAC name, or the logP, etc. |
With a structure on the screen you will get a lot of
calculated data [25]. |
Table 1. Special features and tools in iScienceSearch.
Example:
Search for toxicity
Suppose
we want to learn more about adverse effects of the structure shown in
Figure 1.
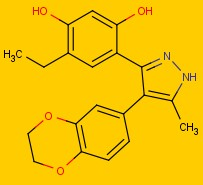
Figure 1: Structure of
4-[4-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-methyl-1H-pyrazol-3-yl]-6-ethylbenzene-1,3-diol
For a comparison, we make a search in SciFinder and iScienceSearch.
Neither SciFinder nor iScienceSearch find something under toxicity (or
adverse effects). In SciFinder, we look for biological studies and find
23 references. In iScienceSearch, we use the profile “Drug Info” and get
an overview as to which database contains information about this
compound, Figure 2.
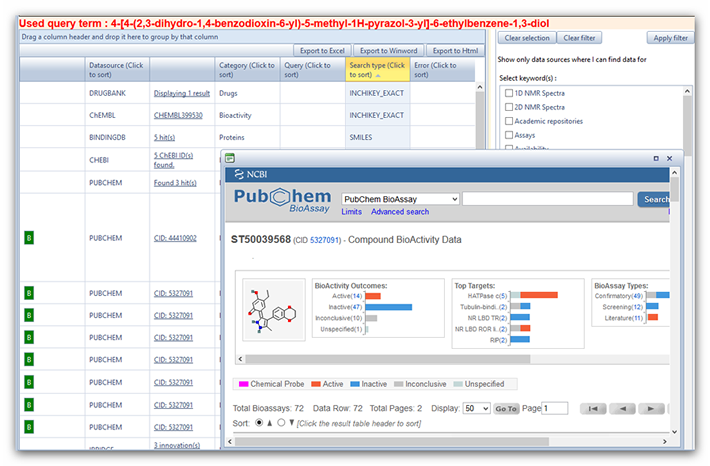
Figure 2: Results of the text search with
4-[4-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-methyl-1H-pyrazol-3-yl]-6-ethylbenzene-1,3-diol
in iScienceSearch. The smaller window shows the result if one clicks on
the green button next to PubChem
PubChem, ChEMBL
[20],
DrugBank [26], etc. have very detailed data, and
very often a good overview of the results. Nobody questions the
usefulness of SciFinder as a literature search tool, but you do not get
an overview as to which database will provide detailed information. In
PubChem, you get a nice overview of articles, and widgets display the
results in iScienceSearch, see BioActivity window in Figure2. You can
select those references first, where the compound is found to be active.
In ChEMBL you get pie charts that help you getting a fast overview of
the activities of a compound.
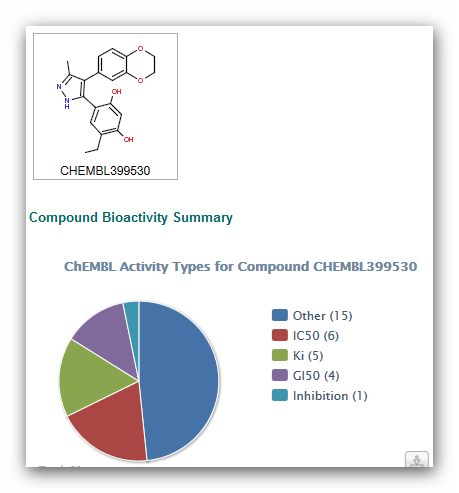
Figure 3: Result in ChEMBL.
Comparison of
iScienceSearch with other databases
No database is as up-to-date as a snapshot of the current status of the
Internet. This means you will not find certain compounds. Try to find in
SciFinder the following structures. We made the search on August 6,
2013, and July 23, 2015.
 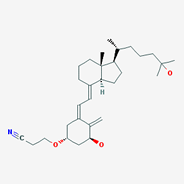 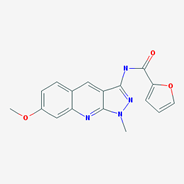
Figure 4: Structures that are not (yet) in SciFinder
Go to
http://www.ncbi.nlm.nih.gov/pcsubstance?cmd=search&term=all%5Bfilt,
and try to find the latest compounds that are recorded in PubChem, and
you will not find a link to it in Google. Even Google takes time to
update its index.
There are currently
(July 24, 2015) 68’417’108 compounds in the PubChem (Compounds)
Database. One can get this count using the url
https://www.ncbi.nlm.nih.gov/pccompound?term=all%5Bfilt%5D.
PubChem is one of the depositor to the ChemSpider database. According to
http://www.chemspider.com/DataSources.aspx
there are currently 10’882’600 reference links to PubChem compounds in
the ChemSpider Database. This means only 16 % of all current PubChem
compounds are referenced in the ChemSpider Database as of today.
The current number of
structure contained in the ChemSpider database mentioned on the
ChemSpider homepage (http://www.chemspider.com/) is 34 Million.
ChemSpider is a depositor to the PubChem database. According to
https://pubchem.ncbi.nlm.nih.gov/sources/sources.cgi
the number of references to compounds in the ChemSpider database is
14’642’781. This means one can only find links to 43% of the ChemSpider
compounds as of today.
Executing an ‘Identical structure’ search in PubChem using the structure
in Figure 1, one only finds a hit for the keto form [27].
Using the same query structure and searching the Drugbank database you
find a hit that reference the enol form [28] in
PubChem. One more reason to use iScienceSearch where you find all the
links.
iScienceSearch only includes free databases. For the ETH (Eidgenössische
Technische Hochschule, Zurich) we have built a “hop-in” button for the
licensed REAXYS system [29] in order to include also
such databases. This means if you have a structure on display you can
search in REAXYS without redrawing or copying the structure.
Biologists do not use SciFinder. They do not have such a database which
collects all abstracts. Biologist are used searching in different
databases. iScienceSearch enables in one search to find answers in many
databases that are interesting for biologists, see list of databases in
the Appendix. Sequence searches are a different story, and you do not do
this in iScienceSearch.
Scifinder and REAXYS are good if you can start with a chemical
structure. They are weak if you start with synonyms. For instance, you
will not find the record in REAXYS starting with “Tracleer”, but only
when you use the less common synonym “Bosentan”. Also, you do not get
the exactly same structure that is in PubChem.
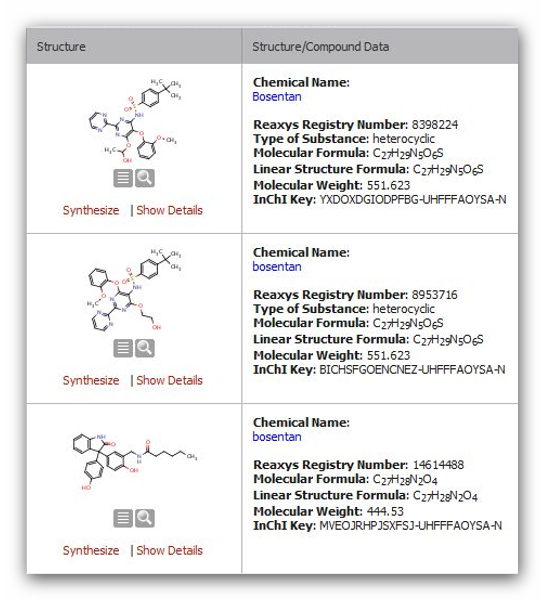
Figure 5: Structures for Bosentan in REAXYS
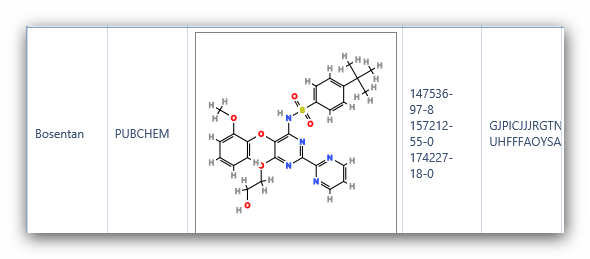
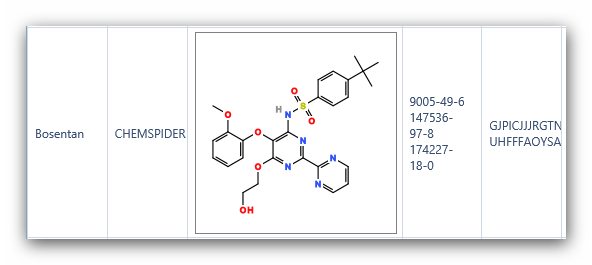
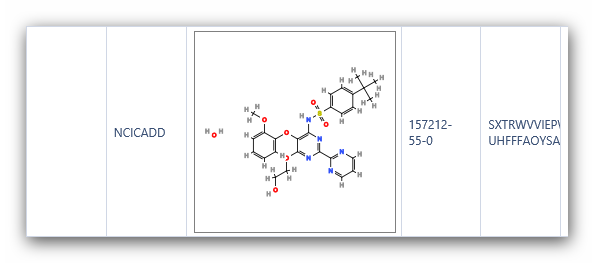
Figure 6:
Structures of Bosentan using “Compare structures” in iScienceSearch.
Have a
look at the InChI key in Figure 8, and it is clear that the structures
in PubChem and REAXYS are different. Checking the InChI key is a
convenient method to quickly differentiate complex structures.
Complex
compounds often have different structures under the same name in
databases. In iScienceSearch we have a possibility to compare the
structures from different databases, pointing immediately to a problem,
alerting the scientist to define his query carefully.
Literature search:
There
are many systems on the Internet, and a user will limit his search to
these sources with which he is familiar. iScienceSearch makes it easy to
search over many sources introducing the user to useful new sources.
Each Internet portal to literature, be it ACS [30],
KonSearch [31], Heidi [32],
etc. has its strength and weakness. Let’s assume a user is fairly
familiar with the different data sources. Let’s assume he is Turkish and
would like to have a quick overview which of the references are in his
mother language. Below is the picture from the query “aspirin toxicity
review” in KonSearch filtered for Turkish documents. Such a filter is a
one click in KonSearch, an option on the right side.
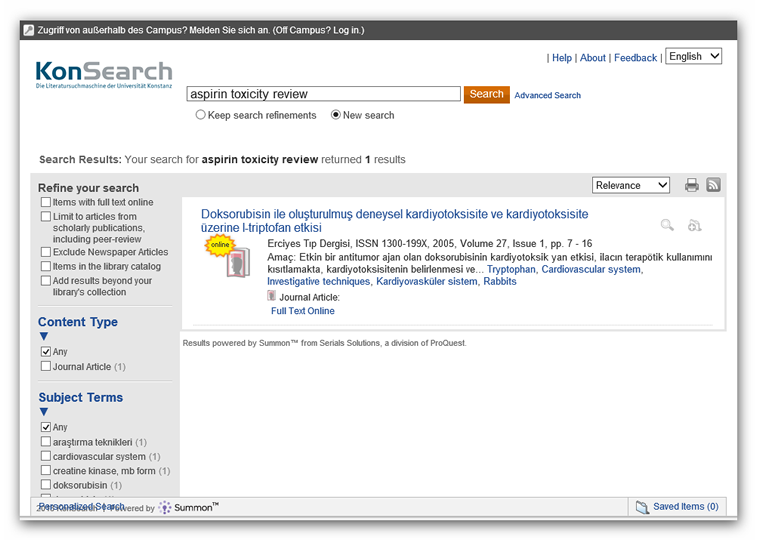
Figure 7:
Example of a search result filtered by language.
Technical Details
iScienceSearch is a meta search engine. A meta-search engine is a
search tool that sends user requests to several other search engines
and/or databases and aggregates the results into a single list or
displays them according to their source [33].
iScienceSearch is an ASP.Net web application hosted under Internet
Information Server (IIS). All searches in iScienceSearch are executed
asynchronously. That allows executing a high number of searches
independent from each other. It also allows interacting with the UI
(user interface) while searches are still executing. This means the
result grid gets populated with links as soon as one of the searches
found a hit. As soon as there are new hits found the result grid gets
updated with those results. Since the UI is not blocked while the search
is executed, the user can open already result links, while searching
goes on in the background. A progress bar shows the search progress in
percentage of completeness. The search can be canceled at any time.
The
chemical drawing tool used in iScienceSearch is JSDraw from Scilligence
Corporation [34]. The editor is written in
JavaScript. That means no Java Plugin need to be installed in the
browser. The only requirement is that the browser has JavaScript
enabled, which is the default setting in all major browsers.
The query extension
(see Extended Search) is using the REST-style version of PUG (Power User
Gateway), a web interface for accessing PubChem data.(
https://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html)
and the Chemical Identifier Resolver from NCI/CADD group (http://cactus.nci.nih.gov/chemical/structure
).
For
predicting chemical properties, (see Additional features) the CAP
(Chemical Activity Predictor) web service provided by NCI/CADD group is
used.
Summary
iScienceSearch provides one user interface to search many databases on
the Internet. The advantage is that one gets a quick overview as to
which source contains relevant information about a compound.
iScienceSearch is unique as an Internet search engine, because it allows
you to search by structure, and not only by text. The extended search
makes it possible to widen the query. With a structure search you find
answers in databases, which for example can only be searched by CAS
Registry Number or text. iScienceSearch provides a short list of links
with the numbers of hits in each source. This makes it easy to pick the
most relevant answers.
Appendix
Data Sources in iScienceSearch.
References and Notes
BIBLIOGRAPHY
|
[1] |
U. J. Heuser,
„Denken, wie das Netz es will,“
Die Zeit, p. 2, 23
September 2010. |
|
[2] |
N. Carr, "Is Google Making Us Stupid?,"
ATLANTIC MAGAZINE,
no. http://www.theatlantic.com/magazine/archive/2008/07/is-google-making-us-stupid/6868/,
July/August 2008. |
|
[3] |
A. Kos and H.-J. Himmler, "CWM Global Search—The Internet Search
Engine for Chemists and Biologists.,"
Future Internet, vol.
2, pp. 635-644, 2010. |
|
[4] |
CAS, "CAS REGISTRY and CAS Registry Number FAQs," 15 8 2013.
[Online]. Available: http://www.cas.org/content/chemical-substances/faqs. |
|
[5] |
"Clove," 23 7 2015. [Online]. Available: https://en.wikipedia.org/wiki/Clove. |
|
[6] |
AKos GmbH, "AKosSamples," 15 8 2013. [Online]. Available:
http://www.akosgmbh.de/AKosSamples/index.html. |
|
[7] |
NCI, "PubMed," 15 8 2013. [Online]. Available: http://www.ncbi.nlm.nih.gov/pubmed. |
|
[8] |
"PubChem -Substance Data Source Information," 15 8 2013.
[Online]. Available: http://pubchem.ncbi.nlm.nih.gov/sources/sources.cgi. |
|
[9] |
CAS, "Chemical Suppliers - CHEMCATS - Find commercially
available chemicals, pricing, and supplier contact information,"
15 8 2013. [Online]. Available: http://www.cas.org/content/chemical-suppliers. |
|
[10] |
A. Hitchcock, "Google's BigTable," 18 10 2005. [Online].
Available: http://andrewhitchcock.org/?post=214. |
|
[11] |
IUPAC, "The IUPAC International Chemical Identifier (InChI)," 15
8 2013. [Online]. Available: http://www.iupac.org/home/publications/e-resources/inchi.html. |
|
[12] |
"Oracle," 22 7 2015. [Online]. Available: http://www.oracle.com/index.html. |
|
[13] |
"MySQL," 22 7 2015. [Online]. Available: http://www.oracle.com/us/products/mysql/overview/index.html. |
|
[14] |
"Crawling & Indexing," 24 7 2015. [Online]. Available: http://www.google.com/insidesearch/howsearchworks/crawling-indexing.html.
[Accessed 24 7 2015]. |
|
[15] |
"Can Google crawl into a database?," 22 7 2015. [Online].
Available: https://www.webmasterworld.com/google/3013128.htm. |
|
[16] |
"eMolecules," 22 7 2015. [Online]. Available: https://www.emolecules.com/. |
|
[17] |
ChemSpider, "Data Sources," 15 8 2013. [Online]. Available:
http://www.chemspider.com/DataSources.aspx. |
|
[18] |
"chemexper," 22 7 2015. [Online]. Available: https://www.chemexper.com/. |
|
[19] |
"SciFinder Content," 22 7 2015. [Online]. Available: http://www.cas.org/products/scifinder/content. |
|
[20] |
"ChEMBL," 22 7 2015. [Online]. Available: https://www.ebi.ac.uk/chembl/. |
|
[21] |
A. Zakharov and M. Sitzmann, "New Web Service: Chemical Activity
Predictor," 15 8 2013. [Online]. Available: http://cactus.nci.nih.gov/blog/?p=1595. |
|
[22] |
ACD / Labs, "ACD / Labs," 15 8 2013. [Online]. Available:
http://www.acdlabs.com/home/. |
|
[23] |
ChemAxon, "chemicalize.org," 17 8 2013. [Online]. Available:
http://www.chemicalize.org/. |
|
[24] |
V. Poroikov, D. Filimonov, T. Gloriozova, A. Lagunin and A.
Lagunin, "Prediction of Biological Activity Spectra for
Substances: in House Applications and Internet Feasibility,"
1998. [Online]. Available: http://www.akosgmbh.de/pass/PASS_Overview.htm. |
|
[25] |
ChemAxon, "chemicalize.org," [Online]. Available: http://www.chemicalize.org/. |
|
[26] |
"DrugBank," 23 7 2015. [Online]. Available: http://www.drugbank.ca/. |
|
[27] |
PubChem Compound, "PubChem Compound - ST50039568 - Compound
Summary (CID 5327091) -keto form," 15 8 2013. [Online].
Available: http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=5327091. |
|
[28] |
PubChem, "Deposited Record (SID 26755036) -enol form," 15 8
2013. [Online]. Available: http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?sid=26755036&viewopt=Deposited. |
|
[29] |
Elsevier, "Reaxys: Chemistry Workflow Solution," 15 8 2013.
[Online]. Available: http://www.elsevier.com/online-tools/reaxys. |
|
[30] |
ACS, "ACS Publications," 15 8 2013. [Online]. Available: http://pubs.acs.org/. |
|
[31] |
University of Konstanz, "KonSearch," 9 8 2012. [Online].
Available: http://www.ub.uni-konstanz.de/digitale-bibliothek/konsearch/. |
|
[32] |
University of Heidelberg, "HEIDI," 2 8 2013. [Online].
Available: http://www.ub.uni-heidelberg.de/helios/kataloge/heidi.html. |
|
[33] |
"From Wikipedia, the free encyclopedia," 17 8 2013. [Online].
Available: http://en.wikipedia.org/wiki/Metasearch_engine. |
|
[34] |
"Scilligence - Software for Life Science," [Online]. Available:
http://www.scilligence.com/web/. [Accessed 24 7 2015]. |
Figures:
Figure
1: Structure of
4-[4-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-methyl-1H-pyrazol-3-yl]-6-ethylbenzene-1,3-diol
Figure 2: Results of the text search with
4-[4-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-methyl-1H-pyrazol-3-yl]-6-ethylbenzene-1,3-diol
in iScienceSearch. The smaller window shows the result if one clicks on
the green button next to PubChem..
Figure 3: Result in ChEMBL.
Figure 4: Structures that are not (yet) in SciFinder
Figure 5: Structures for Bosentan in REAXYS.
Figure 6: Structures of Bosentan using “Compare structures” in
iScienceSearch.
Figure 7: Example of a search result filtered by language.
|
|